
#Gizmo igor pro 7 full#
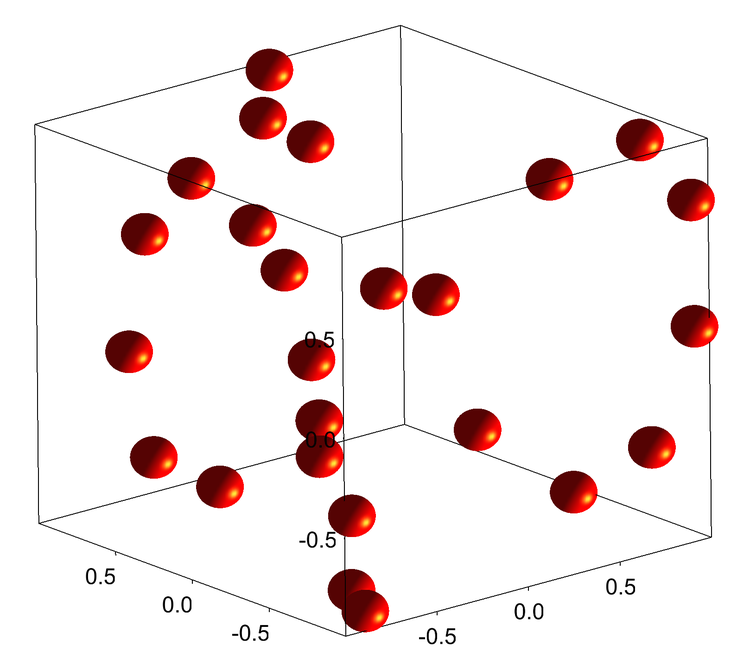
Only the upper-right matrix corner is required, but for coding simplicity the full Adjacency Matrix is constructed in the following function. In the present example the full matrix has N=(60,60), with values either 1 or 0, depending on whether the atoms at the matrix indices are nearest neighbors or not. (If you use a different data set, you should recalculate possibly different bond lengths.) A useful summary of the nearest-neighbor locations is an Adjacency Matrix. There are two possible bond lengths, 1.45331 or 1.36705 Angstrom, for the data file given above. Now we have to figure out which pairs of C atoms are nearest neighbors. Next add to your Procedure window a user utility function that calculates the Cartesian distance between any two C atoms. Combine the three waves 'wx', 'wy', and 'wz' into a new triplet wave 'wxyz'. The included ipf adds a Macro in the main Menu "XYZ waves to XYZ Triplet". this is easily accomplished by using a WaveMetrics utility procedure in your Procedure file: #include It will also be necessary to have a triplet N=(60,3) wave to construct the Gizmo Path object. menu item. Select your text file, skip the first three columns, and load the last three columns into waves named 'wx', 'wy', and 'wz'. Use the Igor Pro 7 Data→Load Waves→Load General Text.
#Gizmo igor pro 7 code#
If you want to exercise the code below and need the Cartesian coordinates for the C60 structure you can use the following link:Ĭenter Atomic Atomic Coordinates (Angstroms)Ĭopy and paste the 60 numerical columns below the section header into a plain-text file, which will have 60 row entries. This molecule is receiving new attention for its potential usage in very small atomic clocks:Ī necessary starting point for the bonds' description is a knowledge of their x, y, and z coordinates.
The structure is highly symmetric and has only two types of C-C bonds and bond lengths, which are either on pentagonal rings, or shared hexagon edges. It is simple because there is only one type of atomic constituent, carbon. The simplest example that interested me is the fullerene molecule C60 (shown above also called buckminsterfullerene, for the famous architect who came up with the "bucky-ball" structure). WaveMetrics provides extensive Help files and Examples on all of Gizmo's aspects if you need to come up to speed. The post is not meant to be a Gizmo tutorial some basic familiarity with Gizmo usage is assumed. I wish to thank AG (WaveMetrics) for suggesting the NaN path-gap idea, and visually enhancing the final Gizmo display. Another useful topic described below is the use of Path waves which contain data for disparate bond segments, separated by NaN rows.
An earlier WaveMetrics example, Molecule.pxp, made with Igor Pro 6, had few molecules, and could not use Igor Pro 7's new Gizmo Path tube-visualization capability.
#Gizmo igor pro 7 manual#
This blog post is meant to show how Igor Pro 7 can be a useful tool for 3D visualization of some molecular bond structures having too many bonds to be input by manual entry.


 0 kommentar(er)
0 kommentar(er)
